Experience
// where I've worked
Senior Computational Biologist
Cultivarium
Cultivarium is a non-profit focused research organization developing open source tools for microbiology. We have developed new computational approaches, designed open molecular toolkits, and engineered novel organisms.
Postdoctoral Researcher
Johns Hopkins School of Public Health
I was a postdoc in Jotham Suez's Lab at JHSPH, studying mechanistic and causative interactions between the gut microbiome and metabolic syndrome.
Computational Biologist
Pattern Ag (merged with EarthOptics)
I designed and developed the soil metagenomic pipelines to detect the most harmful pathogens of major crops in the US.
PhD in Microbiology
University of California, Berkeley
I completed my PhD in the laboratory of Jill Banfield on the evolution, ecology, and genomics of microbial specialized metabolisms, primarily using genome-resolved metagenomics.
B.A. in Biophysics
Johns Hopkins University
Undergraduate studies in biophysics, performing research in the lab of Jocelyne DiRuggiero on metagenomics of extremophiles.
Projects
// a few of the things I've built or discovered
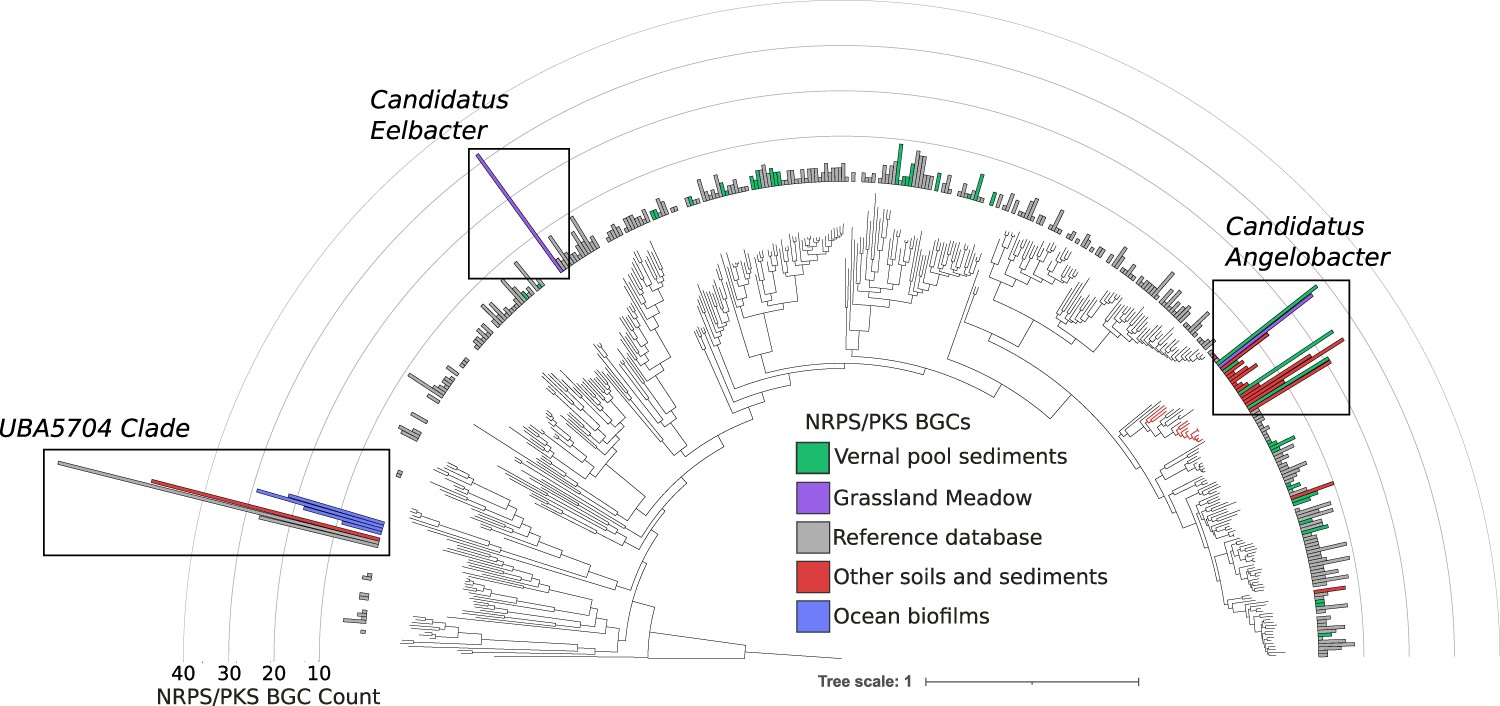
Discovering novel soil microbes
Using genome-resolved metagenomics, I identified deep branching lineages of soil Acidobacteria with genes for production of diverse natural products.

SARS-CoV-2 variant sewage surveillance
Worked on a cross-disciplinary project at UC Berkeley to be one of the first in the world to track exact viral variants using wastewater epidemiology.
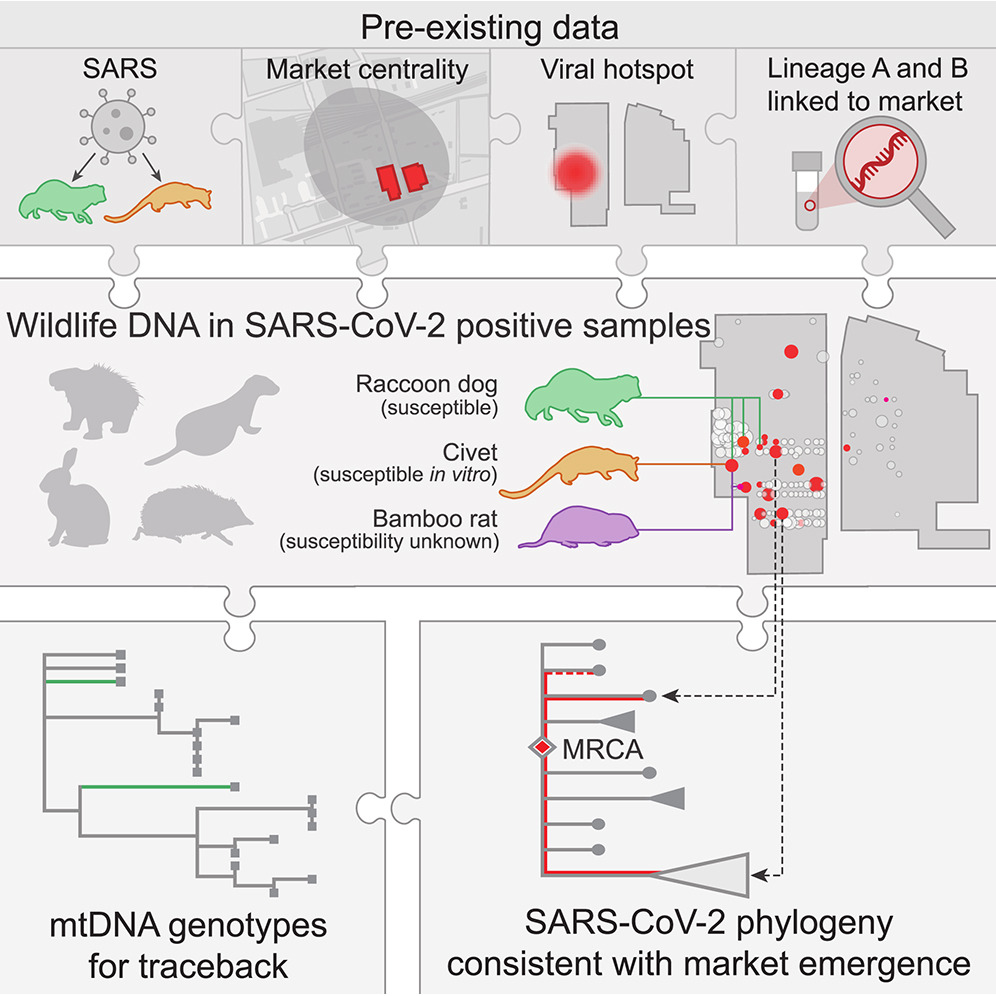
Wildlife origin of SARS-CoV-2
Performed the metagenomic analysis of animal wildlife at the market origin of the COVID-19 pandemic to identify the most plausible intermediate viral hosts.
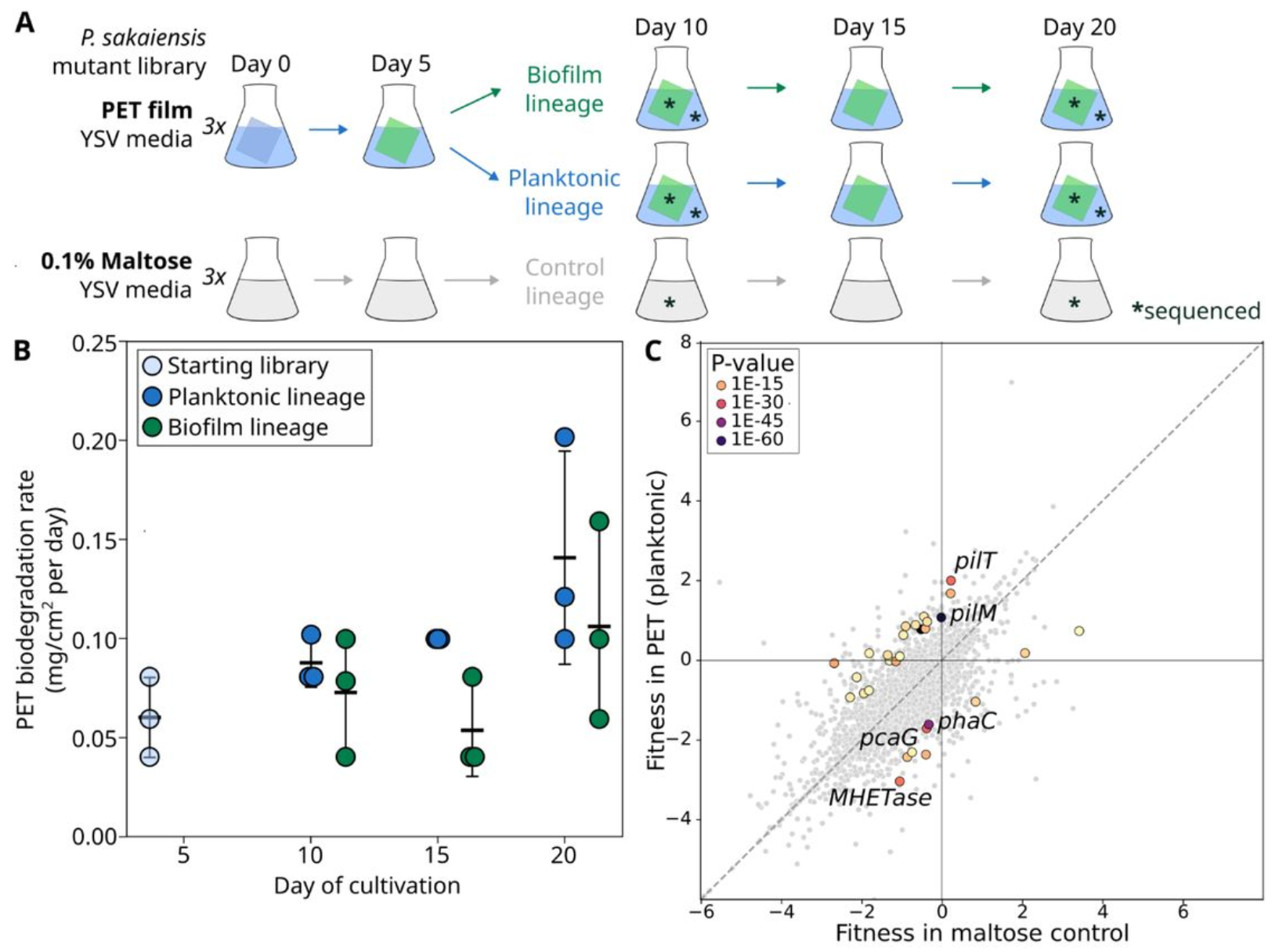
Engineering Ideonella sakaiensis
Published the first genetic tools for the plastic-degrading microbe I. sakaiensis, developing and analyzing a randomly-barcoded mutant library to identify genes involved in plastic degradation.
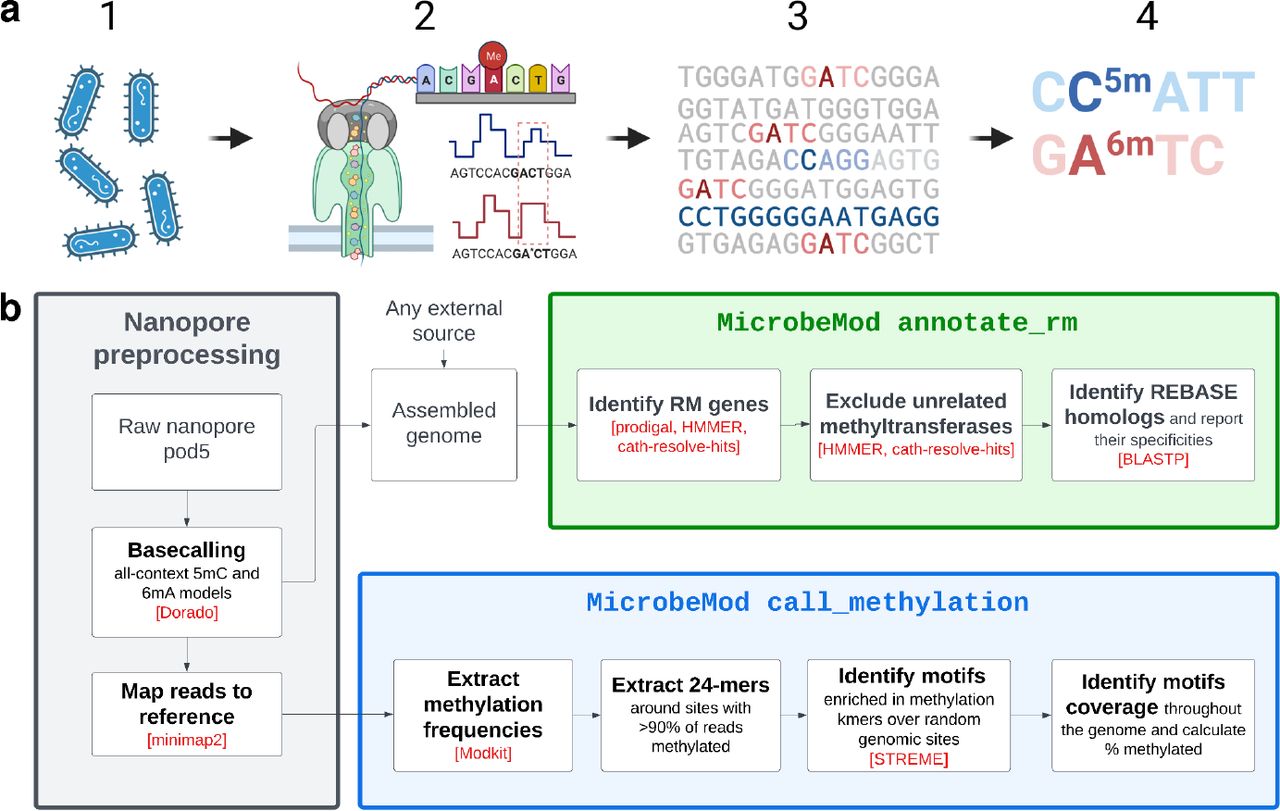
MicrobeMod: prokaryotic methylation detection
Developed MicrobeMod, the earliest software for identifying patterns of three types of prokaryotic methylation using R10 Oxford Nanopore sequencing.
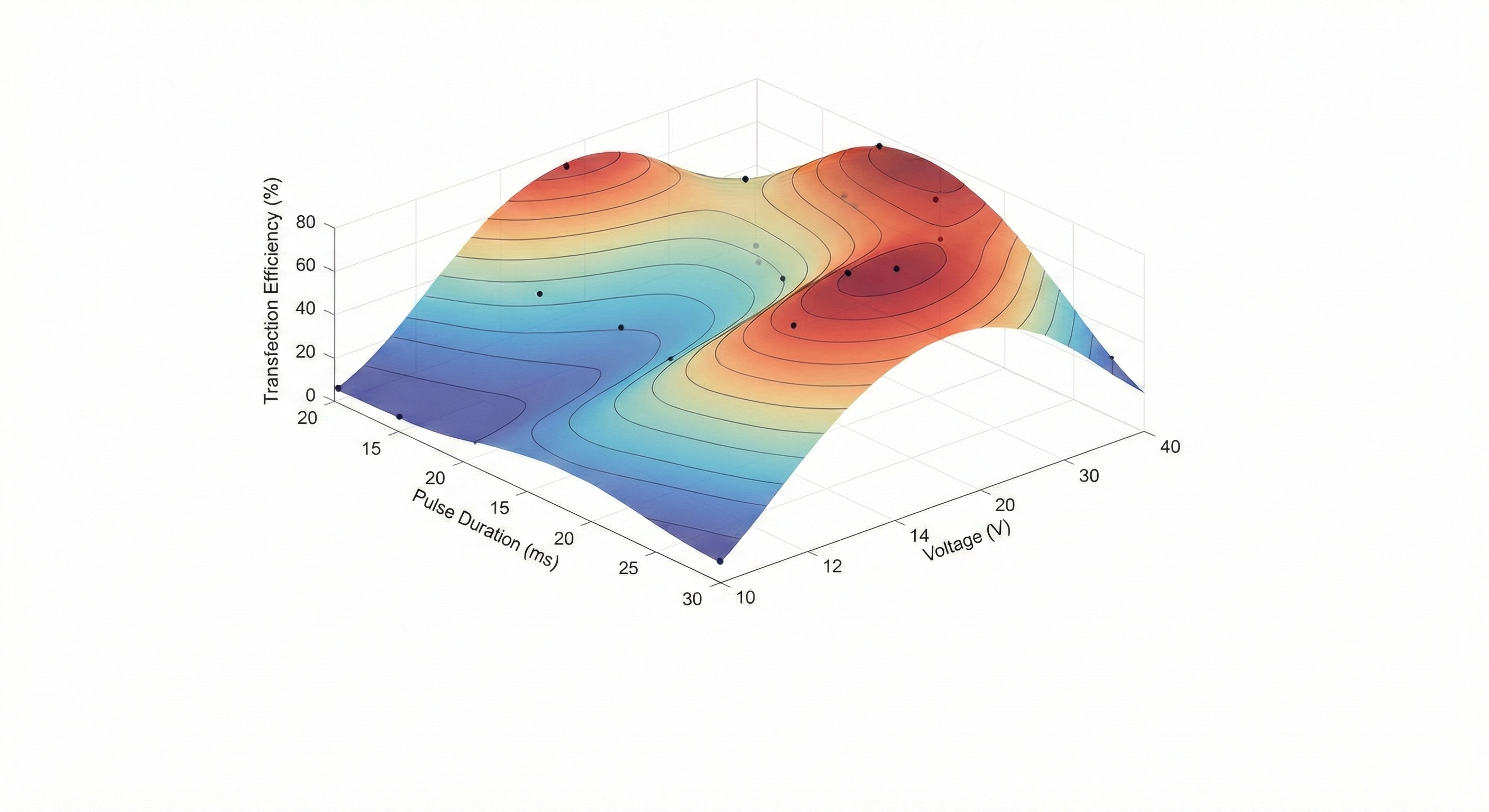
Bayesian optimization for electroporation
Designed a bayesian optimization algorithm to identify optimal microbial electroporation conditions, leading to an improved electroporation protocol for Cupriavidus necator.
Scientific Publications
// 52 papers
Functional genomics in a microbe that degrades and metabolizes PET plastic
Crits-Christoph, Alexander*; Leung, Julia*; Piedra, Felipe-Andrés; Brumwell, Stephanie L; Sajtovich, Victoria A; Abrams, Melanie B; Esmurria, Ariela; Kang, Shinyoung Clair; Mendler, Kerrin; Gilbert, Charlie;
A scalable transposon mutagenesis system for non-model bacteria
Gilbert, Charlie*; Leung, Julia*; Crits-Christoph, Alexander*; Kang, Shinyoung Clair; Esmurria, Ariela; Fenn, Kathrin; Brumwell, Stephanie L; Martin-Moldes, Zaira; Mendler, Kerrin; Barnum, Tyler P;
Genetic tracing of market wildlife and viruses at the epicenter of the COVID-19 pandemic
Crits-Christoph, Alexander; Levy, Joshua I; Pekar, Jonathan E; Goldstein, Stephen A; Singh, Reema; Hensel, Zach; Gangavarapu, Karthik; Rogers, Matthew B; Moshiri, Niema; Garry, Robert F;
MicrobeMod: A computational toolkit for identifying prokaryotic methylation and restriction-modification with nanopore sequencing
Crits-Christoph, Alexander; Kang, Shinyoung Clair; Lee, Henry H; Ostrov, Nili;
A widely distributed genus of soil Acidobacteria genomically enriched in biosynthetic gene clusters
Crits-Christoph, Alexander; Diamond, Spencer; Al-Shayeb, Basem; Valentin-Alvarado, Luis; Banfield, Jillian F;
